Evolving Parallel Computation
Complex Systems, Volume 10, Number 3 (1997)
by Kurt Thearling
and Thomas S. Ray
Abstract
Evolution is applied to the development of parallel digital computer programs. The environment is a shared memory virtual computer containing a population of parallel computer programs. Although the digital computer is a very different environment than the ecology of the Earth, evolution is shown to work effectively in the digital environment and produce significant increases in parallelism.
1 Introduction
The process of evolution by natural selection, until recently, has been known exclusively in carbon-based life on Earth, where it has created stunning diversity and complexity of life forms. However, recent experiments have allowed this process to be extended into the medium of digital computation, producing freely evolving ecological communities of "digital organisms" (self-replicating machine code programs). The challenge becomes understanding and creating the conditions that support diversification and complexity increase in digital evolution in the computer.
Within the realm of carbon-based life, there has been a great complexity increase between the first self-replicating molecules and the large life forms that currently inhabit the Earth. It has been observed that in organic evolution, the bulk of complexity increase occurred in a number of major transitions [1]. The best known and probably largest of these was the "Cambrian explosion of diversity," which was associated with the initial appearance and diversification of macroscopic multi-cellular life on Earth. In the digital domain, analogous transitions might also occur. It is our belief that the transition from serial to digital processes can be compared to the one aspect of the Cambrian explosion, namely the transition from single to multi-cellular organic creatures.
In the work presented here, the first steps in the evolutionary transition from serial to parallel digital processes are described. We show that digital evolution can make use of increasing parallelism to improve the fitness of self-reproducing computer programs. Recent history has shown that parallel computers can be harnessed to efficiently solve complex problems, but optimally implementing such programs can be difficult for human programmers. It is our hope that the results presented here can be used to help generate optimal parallel programs (or critical kernels of parallel programs). Although the problem that is ultimately solved (self-reproduction) is not in itself a terribly useful application, we expect that the general techniques that evolve to optimize the parallelization of this process (e.g., parallel load balancing) will transfer to other applications.
The objective of this work is not to model or simulate organic evolution, but rather to explore the properties of digital evolution. Analogies with organic evolution and life are made, in part to borrow processes that might contribute to complexity increase. However, care must be taken to not force digital evolution to take on forms or processes which mimic organic evolution in ways that are not natural to the digital medium.
In recent years a large body of literature has developed describing the application of digital evolution to various problem spaces. Genetic programming and genetic algorithms [2] are well known examples in this area. Recently the Avida project has explored the evolution of computer programs in more complicated two-dimensional spaces [3]. Our work differs in that we are interested in evolving parallel programs while most other research has focused on optimizing serial (non-parallel) digital processes.
2 Overview of Tierra
The work described here was performed using a software system called Tierra[4]. The Tierra software creates a "virtual computer," a software emulation of a computer. The Tierra computer has a small but otherwise fairly normal instruction set, which is Turing complete and capable of general purpose computation [5]. The virtual operating system (VOS) has some unique process management features that help to support Darwinian evolution. The VOS manages a population of virtual processes, all of which operate on a shared memory area.
Normally, a single replicating program is introduced into the memory. As this program replicates, virtual processes are assigned to the new copies of the program ("daughters"), and each copy of the program is assigned space for its code in the memory area and a single CPU to execute instructions. The VOS provides an equitable sharing of the real CPU time among the virtual processes. When the memory area fills up, the VOS kills old processes to make room for the newborn; thus through time, there will be a turn-over of generations.
The ancestor makes a copy of itself by first determining the memory locations of its beginning and ending, which are indicated by specific instruction patterns in memory. By subtracting the beginning location from the end location the ancestor then determines its length. A new block of memory (of the same size as the ancestor) is then requested for the daughter. The ancestor then enters a loop copying instructions from its memory to the daughter’s new memory. When the copy loop has completed, the ancestor separates from the daughter and the daughter begins executing instructions on its own.
The VOS also introduces noise into the system, in the form of random bit flips in the executable machine code, and in the form of flawed execution of machine instructions. A consequence of this noise is that there is genetic variation among the daughter processes. Due to the turn-over of generations, this generates a process of evolution by natural selection.
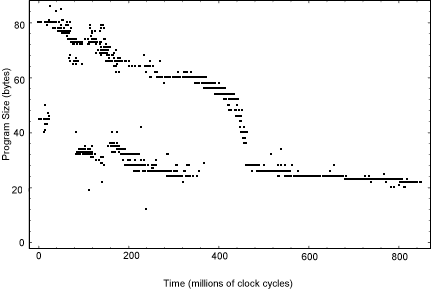
The VOS does not evaluate an explicit fitness measure but implicitly the reproduction time can be thought of as a program’s fitness. Since the reproduction time is proportional to the program size (number of instructions), when all processes are given roughly equal amounts of CPU time, it provides a strong selective pressure for size reduction. A typical run showing the resulting size decrease is illustrated in Figure 1. Initially, there is a bimodal size distribution, reflecting programs near the size of the ancestor (eighty bytes), and much smaller "parasites" which execute the code of neighboring programs. Eventually the fully independent replicating algorithms can reduce in size to as few as twenty-two bytes.
Initial work with the Tierra system demonstrated a great diversity of replicators evolving in a uni-cellular environment [4]. The present report extends this work to explore the feasibility of digital evolution in the context of parallel ("multi-cellular") replicators.

Figure 1. Size optimization of serial self-replicating computer programs. The horizontal axis shows elapsed time in millions of instructions executed by the system while the vertical axis shows genome size in instructions (bytes). Each point indicates the first appearance of a new genotype which crossed a pre-specified abundance threshold of 2% of the population of programs in the memory. |
3 Multi-Cellularity and Parallelism
To allow for the development of parallelism in Tierra, a new ``split’’ instruction was added to the programming language. When a split is executed, an additional CPU is added to the process. This model of developmental parallelism was based on binary cell division as found in living systems and is related to the multi-threaded form of parallelism found in computer science literature [4]. Note that a process with two parallel CPUs is considered to be different than two processes, each with a single CPU (e.g., host and parasite processes). In the former, both CPUs are working on solving the same problem (reproduction of a single copy of the program), while the latter has each CPU working on a different problem (the host copying the host, the parasite copying the parasite, albeit using some of the host’s code to perform the copy).
Each parallel CPU has been given its own independent instruction pointer, which allows for MIMD (Multiple Instruction, Multiple Data) parallelism to develop. In MIMD parallelism, different CPUs can execute different parts of the code. Although MIMD parallelism is the most general (and powerful) form of parallelism, it is often difficult to exploit fully since MIMD programming is a very complicated process.
Each new CPU begins executing at the next instruction following the split. To allow the two parallel threads of execution to differentiate between themselves, at the moment the split occurs, one register is given different values (0 and 1) in the two CPUs. If the processors execute a conditional instruction (e.g., if-zero-then-jump) based on the values in this register, the processors can differentiate themselves from each other by executing different portions of the program.
3.1 Experiments
A large number of experiments in evolving parallel self-reproducing programs have been performed. Over sixty runs were performed generating approximately 312,000 species of multi-cellular Tierra programs. Nearly all runs produced qualitatively similar behavior and this section describes the behavior from a typical multi-cellular simulation. Some of the pitfalls in developing the system, as well as a discussion of how evolution can take advantage of flaws in the system are described by the authors in [7].
As with the original Tierra work [4], we began our experiments with an "ancestor" designed by the authors. The multi-cellular ancestor that was placed into the Tierra memory space started out with a single CPU and then issued a single split instruction. It then proceeded to copy itself using two parallel CPUs in 439 clock cycles. The multi-cellular ancestor operates similar to the previous uni-cellular ancestor, except that the ancestor’s two parallel processors each copy different blocks of memory to the daughter. One processor copies the first half of the ancestor’s memory while the other processor copies the second half.
The question then was whether evolution would be able to improve the efficiency of the self-reproducing program by increasing the use of parallelism. Since multiple CPUs are now possible for a single self-reproducing program, the relationship between size and reproduction time is no longer directly inferable from the size. A long program with many parallel CPUs might very well reproduce faster than a short program with few parallel CPUs.
The graph in Figure 2 shows the evolution of reproduction time for a typical multi-cellular simulation. The run is initialized with the ancestor program which quickly fills up the simulated computer’s memory. In nearly all of the multi-cellular Tierra simulations that were run, two distinct evolutionary phases can be observed after the initial population develops. In the first phase, the evolutionary process makes use of only serial program optimization techniques to reduce program size. Once the serial optimizations have run their course, the second phase begins and evolution introduces additional parallelism to decrease reproduction time.
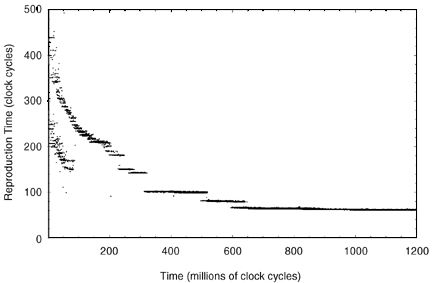
Figure 2. Optimization of reproduction time in parallel programs. Horizontal axis same as Fig. 1, vertical axis number of clock cycles required to reproduce. Over 14,000 unique program species exceeded the abundance threshold and were saved during a simulation totaling 1.2 billion instructions. |
During the first evolutionary phase shown in Figure 2, two bands of program genotypes (instruction sequences) can be seen. The upper band corresponds to the fully self-reproductive programs while the lower band corresponds to parasites. During the 200 million clock cycles associated with phase one, the programs gradually improved their reproduction speed by reducing their size, but there was no increase in parallelism detected, beyond the two CPUs built into the ancestor.
Following the completion of the first evolutionary phase, sharp decreases in reproduction time (e.g., 30% in Figure 2) are usually observed. This optimization is achieved through additional parallelism, corresponding to an increase from two to four CPUs per program. In the graph of program size versus time (Figure 3, from the same run as shown in Figure 2), this change is even more noticeable since the newly evolving programs increased in size from 36 to 44 bytes. The larger but more parallel programs take over the population.
Once additional parallelism evolves, it becomes the dominant form of improvement in reproduction efficiency. At about 500 million clock cycles programs with eight CPUs can be seen. About 100 million clock cycles later evolution pushes to sixteen CPUs (which was the simulator’s limit in this run).
Other runs with higher limits (between 64 and 256 CPUs) have evolved programs that were able to use 32 parallel CPUs. It appears that 32 CPUs is the limit of usefulness of parallelism for the programs in their current form. In fact, some runs that evolved 32 CPU parallelism sometimes oscillated between 16 and 32 CPUs after being allowed to run for extended periods. This is due to the fact that 16 CPU size 60 programs require almost exactly the same reproduction time as 32 CPU size 70 programs. At this point in the evolutionary process, for 16 and 32 CPUs the fitness differential is negligible.
3.2 Evolutionary Tricks
One noticeable characteristic that can be seen in the size versus time graph (Figure 3) is that when there are increases in parallelism, the first programs with the increased parallelism generally have a size that is a multiple of the number of CPUs. These programs usually contain some instructions that have no effect (e.g., incrementing an unused register). Although these instructions do not affect the operation of the program, they pad its size to a multiple of the number of CPUs so that the workload can easily be evenly distributed among the parallel CPUs.
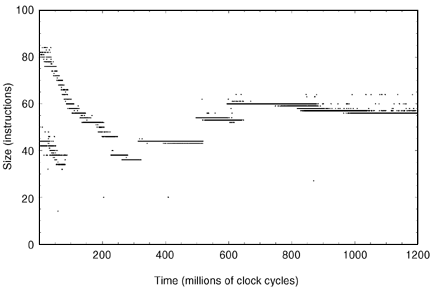
Figure 3. Size optimization of parallel programs. Axes the same as in Figure 1.
After each increase in parallelism, evolutionary optimization begins to remove the unnecessary instructions to make the programs shorter. This reduction in size is usually interrupted by additional increases in parallelism, until the programs have reached their CPU limit. In that case, the only optimization that can be performed is size reduction. Notice that the size reduction at the end (16 CPUs) lasts significantly longer than the size reductions for earlier increases in parallelism.
Digital evolution produced some interesting behavior when optimizing multi-cellular Tierra programs. For example, it is difficult to equally distribute the workload of copying 60 instructions by 16 processors. One simple solution would be for each CPU to copy four successive instructions, with the last CPU copying four extra instructions beyond the end of the program. This approach would usually incur a penalty since the memory beyond the end of the daughter program is generally owned and write protected by another process. Even though this algorithm would incur a penalty, it would be able to reproduce quite effectively.
Evolution was able to choose a slightly more complicated approach to this workload distribution problem. Figure 4 shows the decomposition of the problem of self-reproduction for a 60 instruction, 16 CPU program. The execution of the program hierarchically decomposes the problem, first starting with two CPUs (each copying 30 instructions). The increase in parallelism to four CPUs has each CPU copying a separate 15 instruction portion of the program.
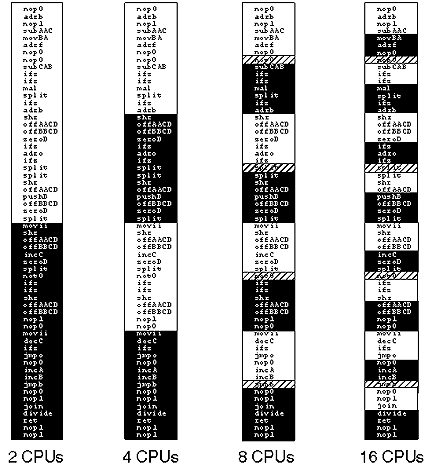
Figure 4. Workload distribution during increasing parallelism for a 60 instruction self-replicating program. |
When the parallelism increases to eight CPUs, the decomposition adds a slight twist. At this point each CPU copies eight instructions, but the four pairs of CPUs each overlap one of their instructions so that the extra instructions are written to the daughter twice. This decomposition proceeds in a similar fashion for the increase to 16 CPUs. Evolution has found a very elegant solution to the problem of workload distribution among parallel processors.
4 Conclusion
The evolved parallel programs observed in this study did not exhibit full MIMD parallelism. Although each CPU had its own instruction pointer, the simplicity involved with copying a block of data from one location to another did not require that different CPUs perform significantly different tasks. Each CPU is able to execute the same instructions, with the only difference being that different CPUs copy different parts of a program. The next step in this work is to allow programs to exist in a much larger, networked computer environment [8]. It is hoped that this will challenge evolution with even more complex problems, leading to the evolution of differentiated, MIMD parallel and distributed software.
References
[1] J. Maynard Smith and E. Szathmáry, The Major Transitions in Evolution (Freeman, Oxford, 1995).
[2] M. Mitchell, An Introduction to Genetic Algorithms (MIT Press, 1996); J. Koza, Genetic Programming (MIT Press, 1993).
[3] C. Adami and C.T. Brown, in Artificial Life IV, R. Brooks and P. Maes, Eds., (MIT Press, Cambridge, MA, 1994).
[4] T.S. Ray, in Artificial Life II, Santa Fe Institute Studies in the Sciences of Complexity, vol. X, C. Langton, C. Taylor, J. D. Farmer, & S. Rasmussen, Eds. (Addison-Wesley, Redwood City, CA, 1991.
[5] C.C. Maley, M.S. thesis, New College, Oxford University (1993).
[6] Multithreaded Programming Guide (Sun Microsystems, Mountain View, CA, 1994).
[8] T.S. Ray, Technical Report TR-H-133 (ATR Laboratories, 1995).