 The creature occupies a space of 960 bytes, which
includes 320 bytes for the genome, a 512 byte area for data,
and 128 bytes of unused space.
The creature occupies a space of 960 bytes, which
includes 320 bytes for the genome, a 512 byte area for data,
and 128 bytes of unused space.
Based on the enhancements to the instruction set described above, in April 1996, the first fully network-able ancestor was designed and implemented. Over the following year, the algorithm was refined through experience. What follows is a description of the algorithm of the network ancestor, whose assembler code is available at: http://www.isd.atr.co.jp/~ray/tierra/netreport/0960aaa.tie.July.
The overall structure of the network ancestor can be seen in Figure 1.
 The creature occupies a space of 960 bytes, which
includes 320 bytes for the genome, a 512 byte area for data,
and 128 bytes of unused space.
The creature occupies a space of 960 bytes, which
includes 320 bytes for the genome, a 512 byte area for data,
and 128 bytes of unused space.
The structure of the genome can be seen in Figure 2.
 The genome is defined as that code
that is copied from mother to daughter, and is therefore
heritable. The genome is considered to contain the following
genes:
sel self-examination,
dif differentiation,
rep reproduction,
cop data copy,
dev development, and
sen sensory. And there is a 27 byte unexpressed area at the
end of the genome.
The genome is defined as that code
that is copied from mother to daughter, and is therefore
heritable. The genome is considered to contain the following
genes:
sel self-examination,
dif differentiation,
rep reproduction,
cop data copy,
dev development, and
sen sensory. And there is a 27 byte unexpressed area at the
end of the genome.
The sel gene is expressed only by the undifferentiated initial cell. The dif gene is expressed only by the initial cell, and the undifferentiated products of its first division. Execution of the dif gene causes the first two cells to differentiate into reproductive and sensory cells (by means of a conditional jump to the rep or sen genes). The rep gene is expressed only by the reproductive tissue, and the sen gene is expressed only by the sensory tissue. The cop and dev genes are expressed by both reproductive and sensory tissues, through a system of gene promotion (procedure calls).
The deterministic cell lineage of the network ancestor is illustrated
in Figure 3.
 After differentiation into reproductive and sensory
cells, the reproductive cell divides into a tissue of two cells
which share the work of copying the genome from mother to
daughter. The sensory cell undergoes three successive divisions
producing a tissue of eight sensory cells. These eight cells
work in parallel to gather and analyze TPing sensory data.
After differentiation into reproductive and sensory
cells, the reproductive cell divides into a tissue of two cells
which share the work of copying the genome from mother to
daughter. The sensory cell undergoes three successive divisions
producing a tissue of eight sensory cells. These eight cells
work in parallel to gather and analyze TPing sensory data.
The two reproductive cells spend a long time copying the genome, which is completed 1030 (parallel) instructions after the start of the network ancestor program. At 1035 instructions, one of the two reproductive cells dies (halts), leaving the single remaining reproductive cell to execute the divide instruction, which spawns the daughter as an independent process. After spawning the daughter, the reproductive cell divides again into two cells, and repeats the process in an infinite loop.
Just before spawning the daughter, the reproductive cell reads an IP address (the address of a machine participating in the Tierra network) from a location near the beginning of the 512 byte data area. At birth, the daughter is sent to the machine at this IP address, and the daughter genome is erased from the local machine.
While the reproductive tissue is engaged in the long process of
copying the genome, the sensory tissue is gathering and reducing
sensory data, Figure 4.
 The 512 byte data area is used to hold eight
sixty-four byte TPing data structures, gathered from eight different machines
on the Tierra network. The eight TPing data structures are subjected
to three rounds of data reduction, in which pairwise comparisons
of Speed/NumCells are made. If the data on the right has a
higher ratio than the data on the left, the data on the right is
copied over the data on the left. At the end of the data reduction,
the data corresponding to the machine with the highest Speed/NumCells
ratio will be located in the first of the eight blocks of data.
This is where the reproductive cell will look for the IP address to which
the daughter will be sent.
The 512 byte data area is used to hold eight
sixty-four byte TPing data structures, gathered from eight different machines
on the Tierra network. The eight TPing data structures are subjected
to three rounds of data reduction, in which pairwise comparisons
of Speed/NumCells are made. If the data on the right has a
higher ratio than the data on the left, the data on the right is
copied over the data on the left. At the end of the data reduction,
the data corresponding to the machine with the highest Speed/NumCells
ratio will be located in the first of the eight blocks of data.
This is where the reproductive cell will look for the IP address to which
the daughter will be sent.
The developmental behavior of the sensory cells is illustrated in Figure 5.
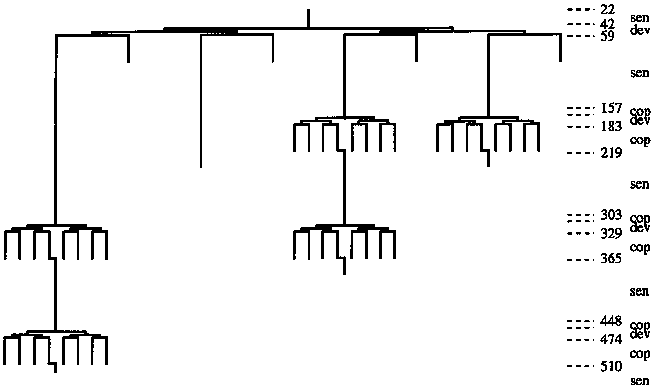 Initially, each of the eight sensory cells reads one TPing data
structure into its sixty-four byte memory block. Then half of the eight
sensory cells die (halt). The remaining four cells conduct the
first round of pair-wise comparisons. If the data on the right
is better than the data on the left, the corresponding sensory
cell will promote the cop gene, splitting into eight
threads to copy the data from right to left. After the data is
copied, seven of the eight cells die. Next, two of the four
remaining sensory cells die, and the two remaining cells conduct
the second round of pair-wise comparisons. Next, one of the two
sensory cells dies, and the single remaining cell conducts the
third and last pair-wise comparison.
Initially, each of the eight sensory cells reads one TPing data
structure into its sixty-four byte memory block. Then half of the eight
sensory cells die (halt). The remaining four cells conduct the
first round of pair-wise comparisons. If the data on the right
is better than the data on the left, the corresponding sensory
cell will promote the cop gene, splitting into eight
threads to copy the data from right to left. After the data is
copied, seven of the eight cells die. Next, two of the four
remaining sensory cells die, and the two remaining cells conduct
the second round of pair-wise comparisons. Next, one of the two
sensory cells dies, and the single remaining cell conducts the
third and last pair-wise comparison.
This process of data collection and reduction takes about 510 (parallel) instructions to complete, thus, in the time that it takes the reproductive tissue to copy the genome, the sensory system is able to complete two sensory cycles, resulting in the gathering and analysis of TPing data from fifteen nodes (after the first sensory cycle, the sensory cell using the left-most sixty-four byte data block, does not read a TPing data structure, so that the best data from the previous sensory cycle can be remembered).